1.4 Genome-wide CRISPR–Cas9 screening reveals ubiquitous T cell cancer targeting via the monomorphic MHC class I-related protein MR1
DOI: https://doi.org/10.1038/s41590-019-0578-8
Michael D. Crowther, Garry Dolton, Mateusz Legut, Marine E. Caillaud, Angharad Lloyd, Meriem Attaf, Sarah A. E. Galloway, Cristina Rius, Colin P. Farrell, Barbara Szomolay, Ann Ager, Alan L. Parker, Anna Fuller, Marco Donia, James McCluskey, Jamie Rossjohn, Inge Marie Svane, John D. Phillips & Andrew K. Sewell
Year: 2020
Affiliations Cardiff University
Fig. 1: MC.7.G5 recognizes multiple cancer types through an HLA-independent mechanism.
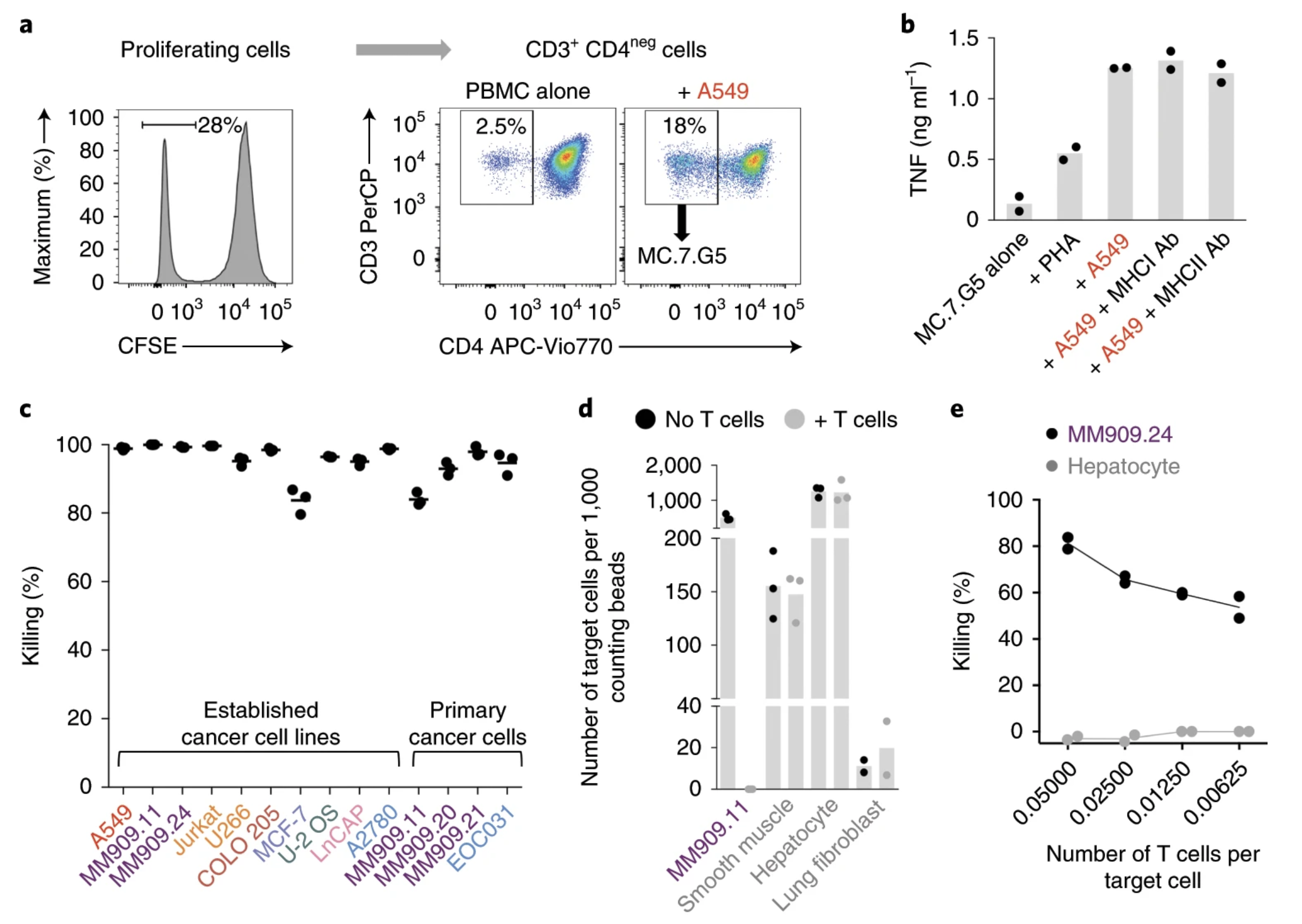
Fig. 2: Whole-genome CRISPR–Cas9 library screening reveals MR1 as the candidate target of MC.7.G5.
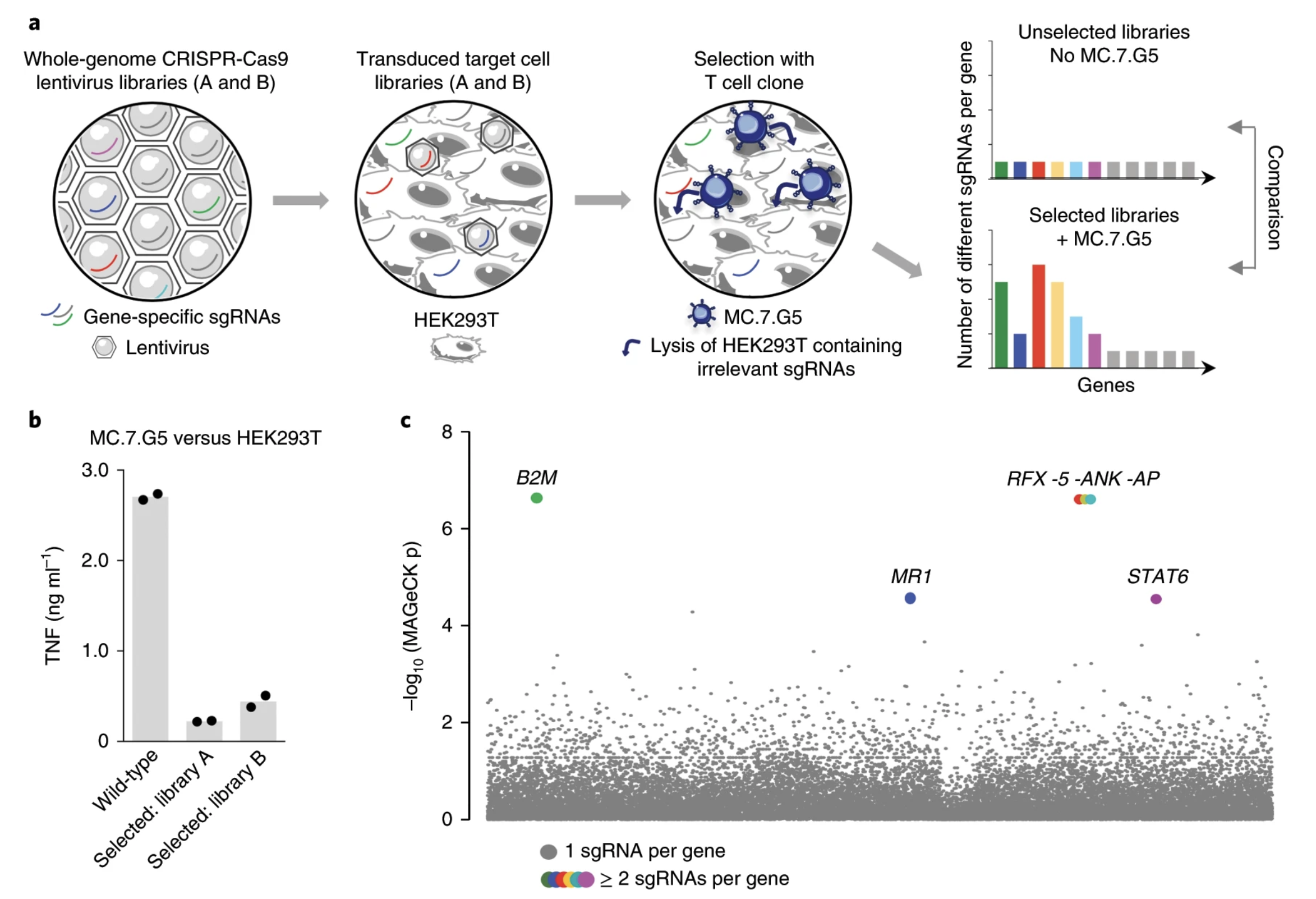
Fig. 3: MR1 is the cancer cell-expressed target of MC.7.G5.
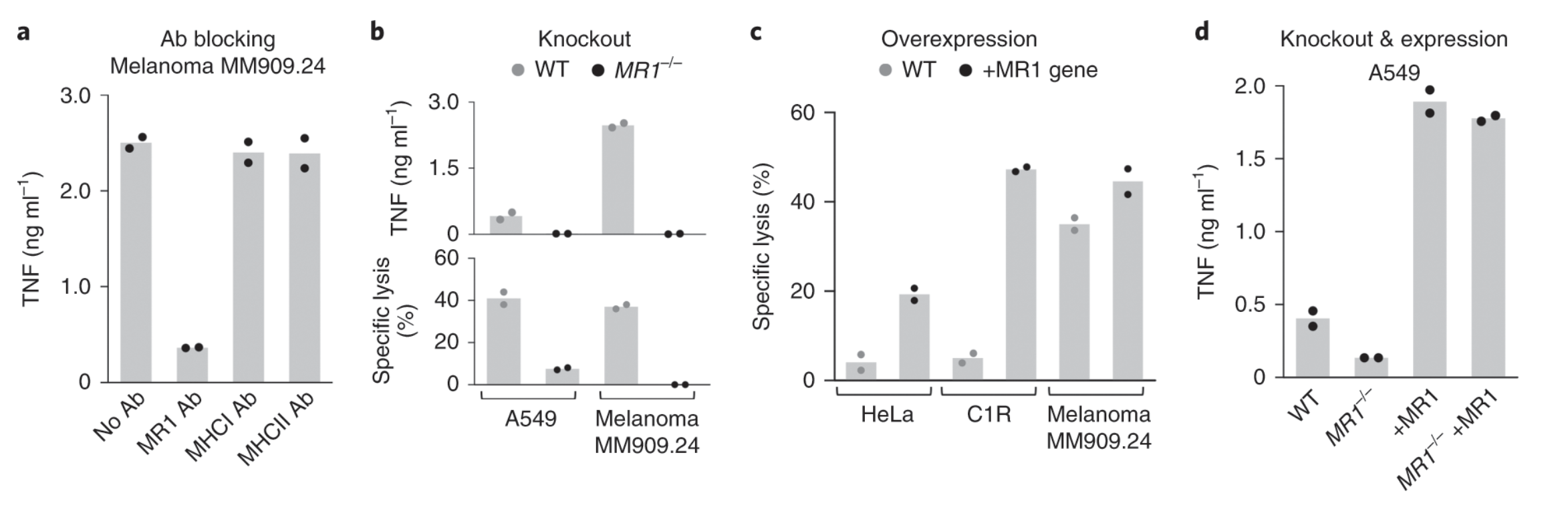
Fig. 4: MC.7.G5 does not recognize MR1 by known mechanisms.
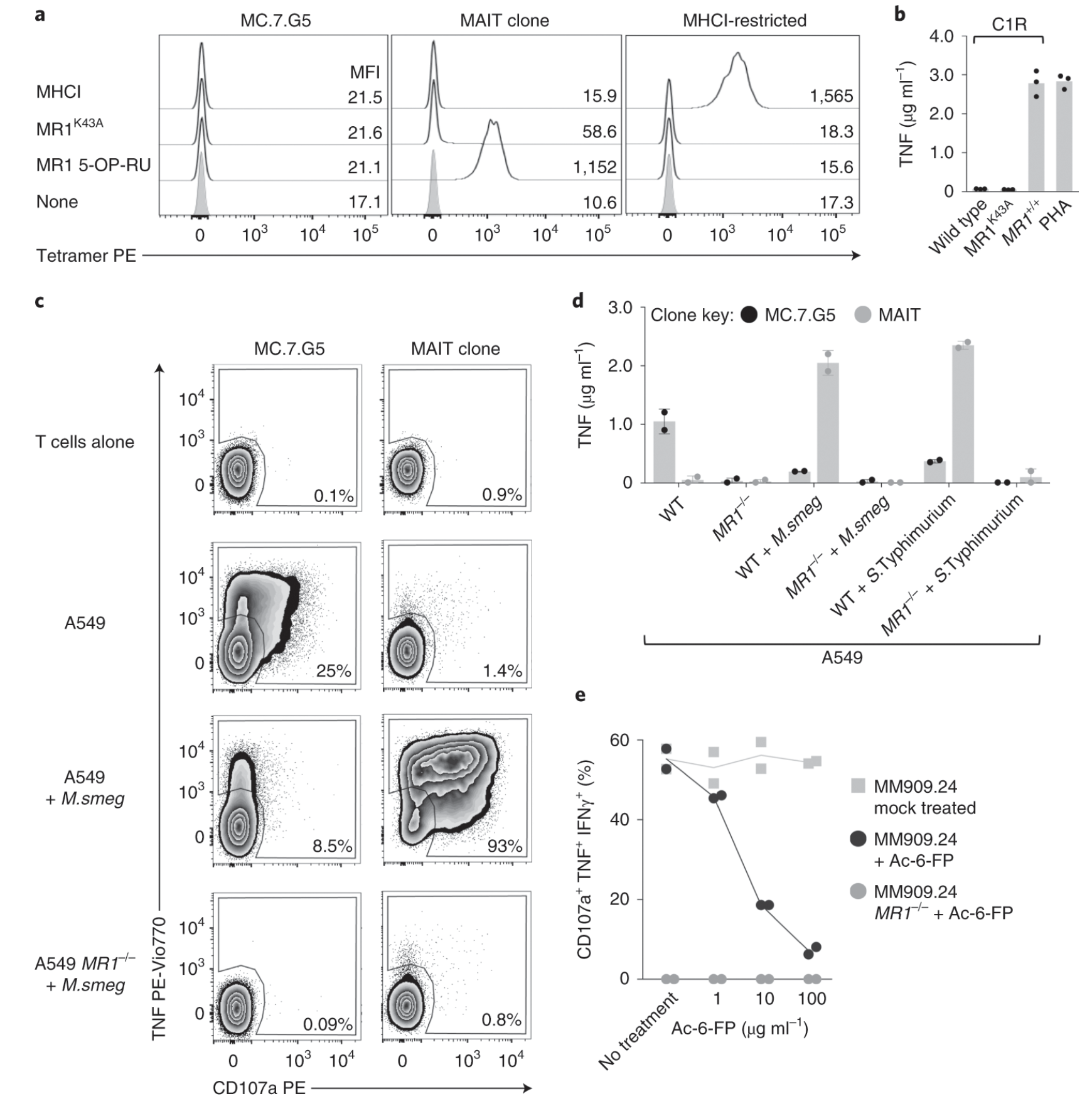
Fig. 5: MC.7.G5 does not recognize healthy cells.
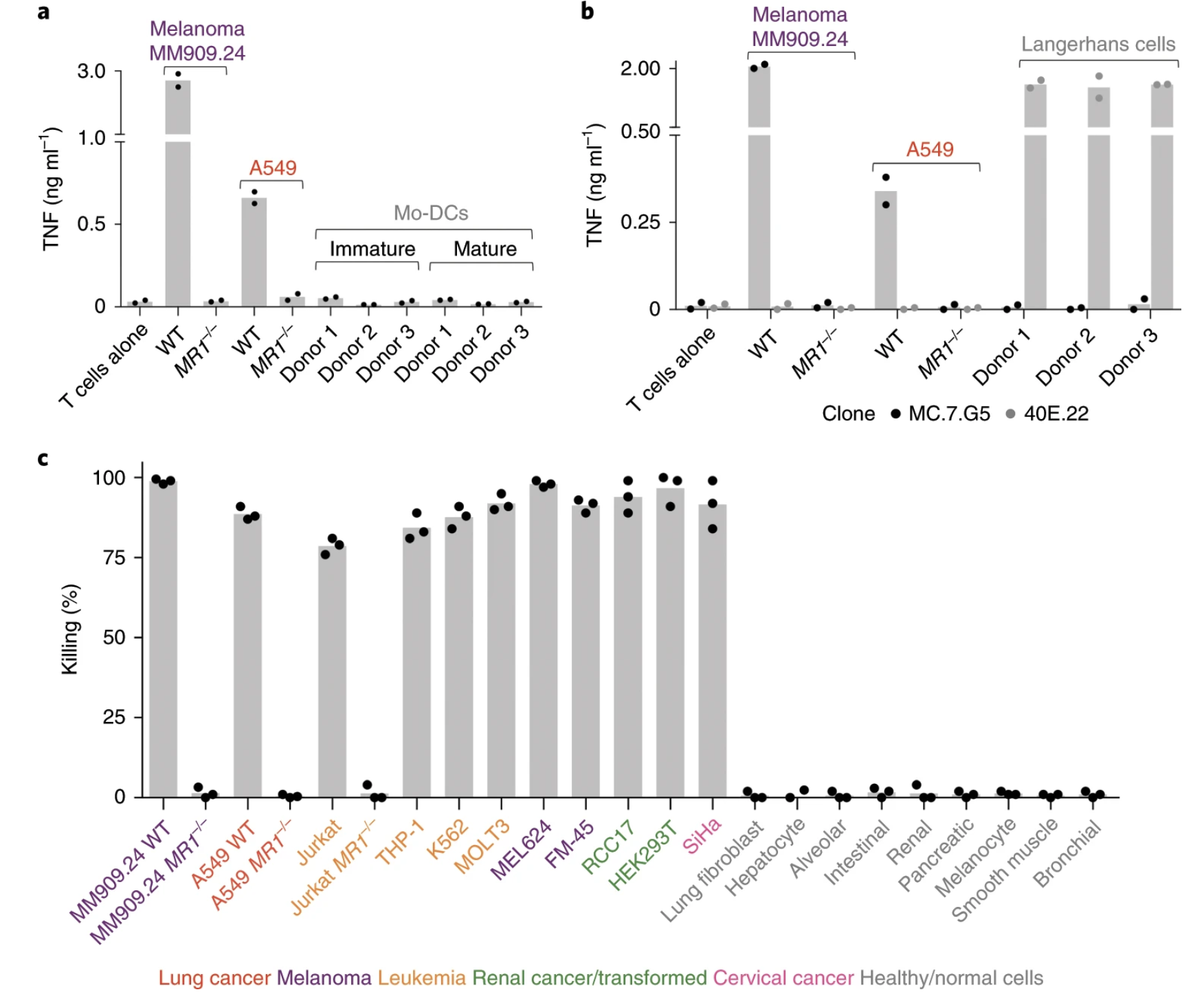
Fig. 6: MC.7.G5 remained inert to activated, stressed or infected healthy cells.
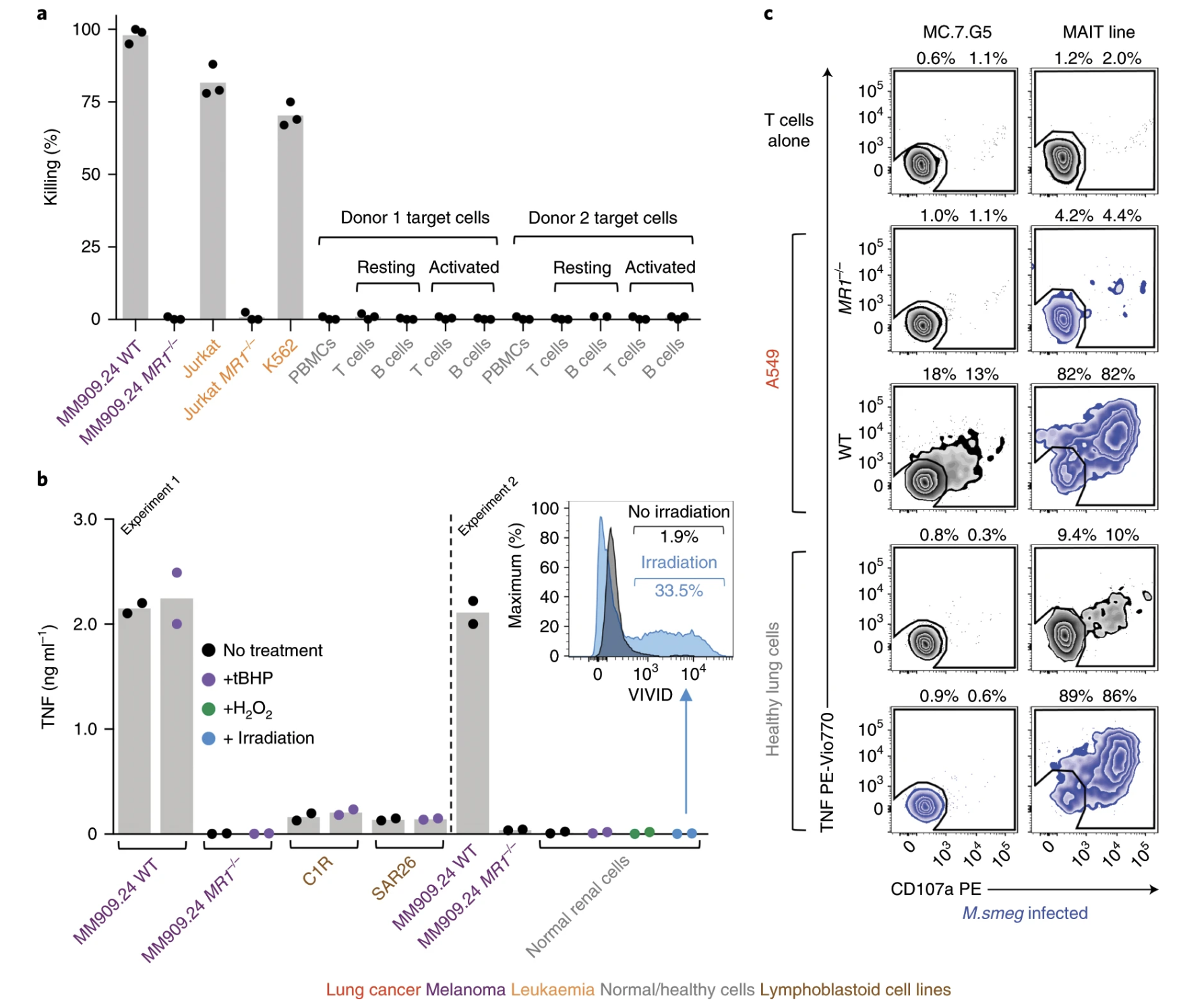
Fig. 7: MC.7.G5 mediates in vivo regression of leukemia and prolongs the survival of mice.
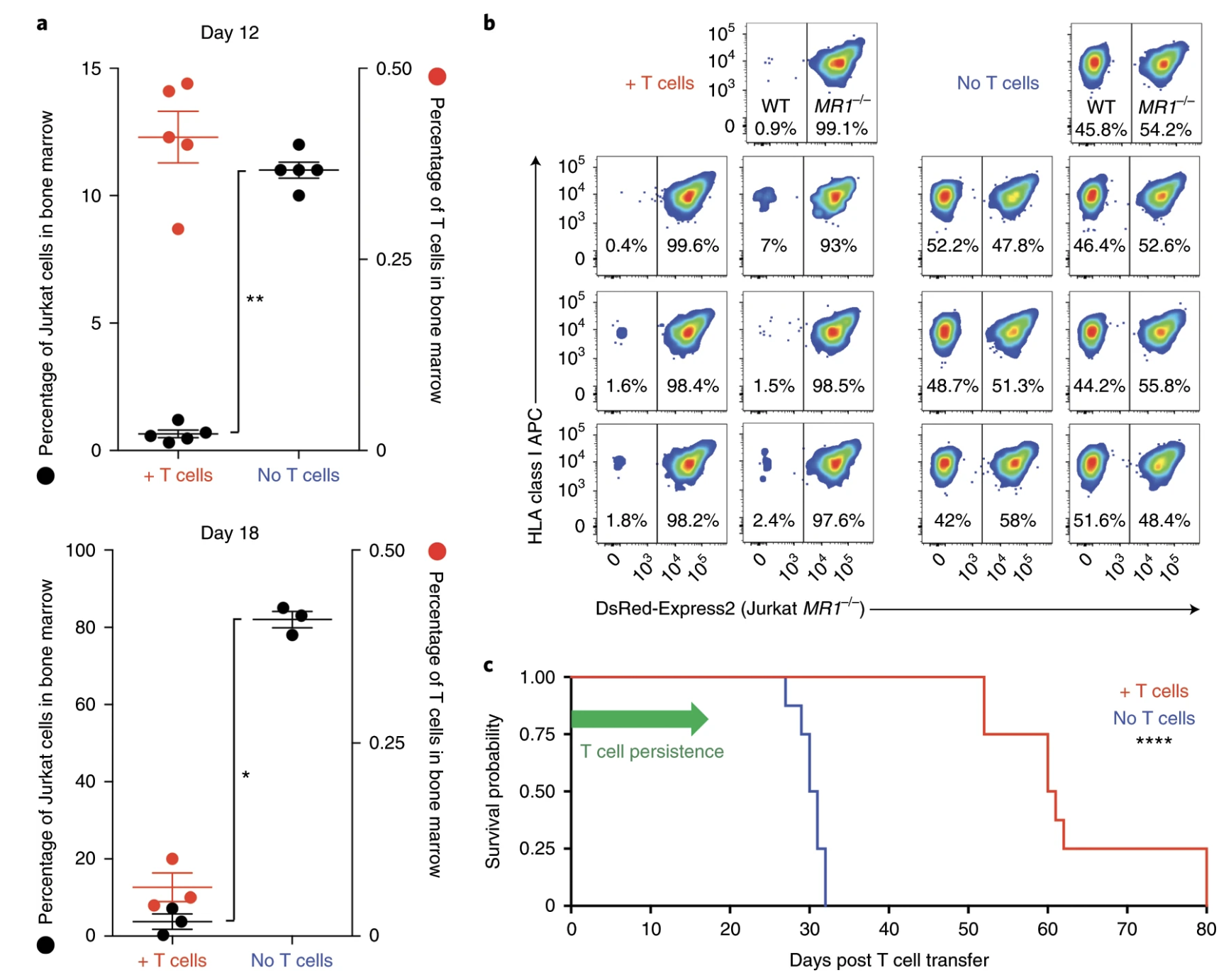
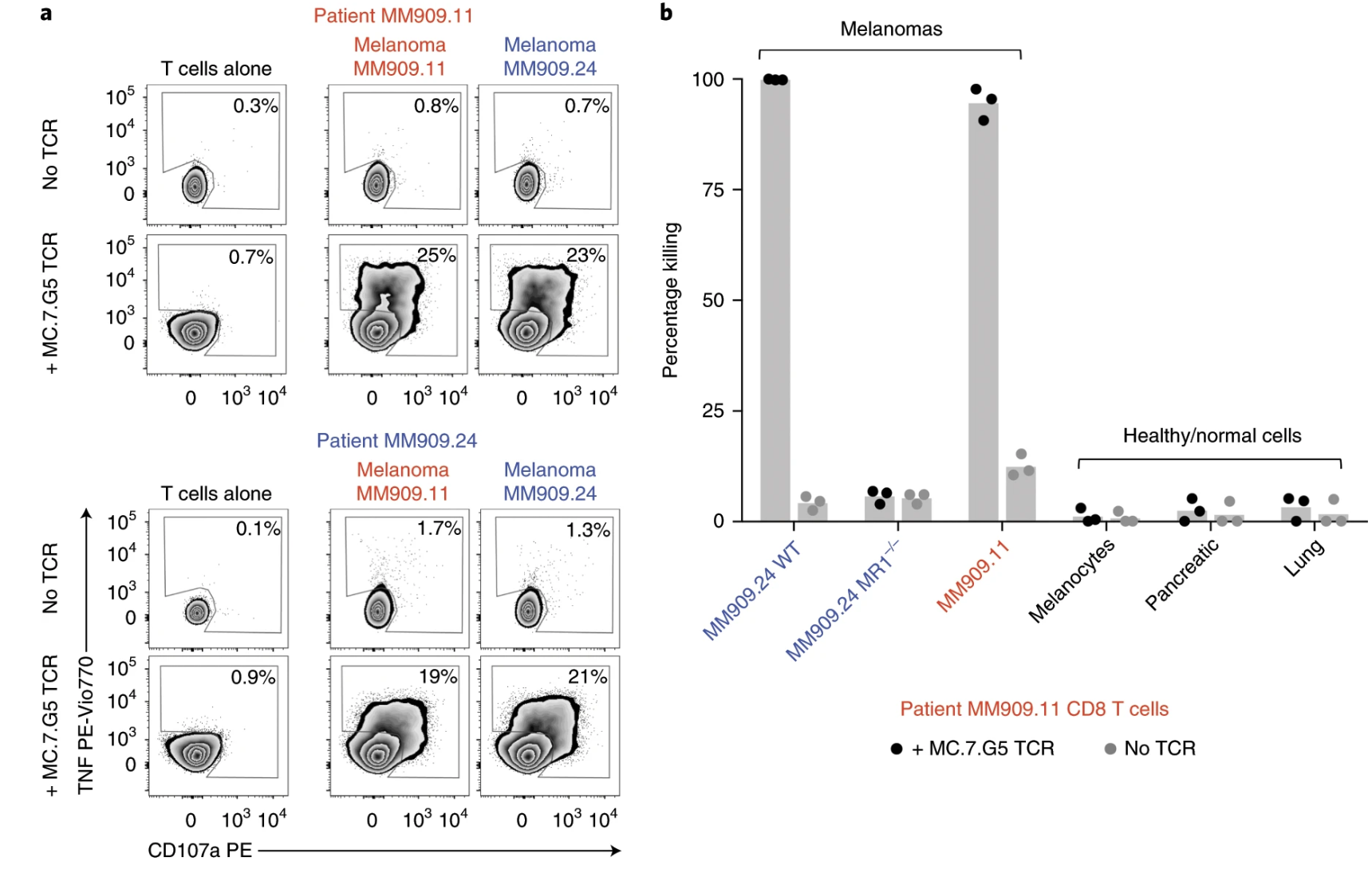
Fig. 8: Transfer of the MC.7.G5 T cell receptor redirects patient T cells to recognize autologous melanoma.
Summary, novelty, and main findings:
Ligands of unconventional T cell receptors are unknown. Previous studies established some ligands of unconventional T cell receptors as biosynthetic intermediates. T cell recognition of metabolic intermediates was strengthened through the discovery of mucosal-associated invariant T (MAIT) cells recognizing MHC class 1-related protein (MR1) bound to microbial metabolites. This paper studies non-MAIT cell non-bacterial cell recognition of MR1 that results in the killing of cancer cells. A clone of T cells (MC.7.G5) that proliferated in co-culture of A549 cancer cells and peripheral blood mononuclear cells (PBMC) were accessed for killing independent of MHC recognition (Figure 1). A CRISPR screen was utilized to identify MR1 as a key gene in MC.7.G5-HEK293T binding (Figure 2 and 3). MC.7.G5 T cells did not recognize microbial related MR1s or MR1-bound ligands and exhibited specificity to cancer cells (Figure 3 and 4). MR1 led to various cancer cell lysis but not normal cells and stress or bacterial infection did not lead to MC.7.G5 activation (Figure 5 and 6). Adopted transfer of MC.7.G5 T cells targeted and decreased cell burden of Jurkat cells that were engrafted into mice (Figure 7). Melanoma patients derived T cells were transduced with MC.7.G5 which led to the killing of autologous and non-autologous melanomas (Figure 8).
Most important figures:
The most important figures for this paper were figures 6, 7, and 8. Figure 6 continues the proof that MC.7.G5 was only activated in the presence of cancer cells and not due to any other non-specific interaction (bacterial or stress). Figure 7 moves MC.7.G5 into an in vivo mouse model to identify a reduction of cancer cell burden compared to the control. Lastly, Figure 8 indicates through an ex vivo human model to transduce MC.7.G5 T cells to kill autologous cancer from patients.
Holes of the paper:
I think for Figure 7 it was cool that they pointed out that there was a reduction of Jurkat cell burden, but it would have been more powerful to point at different expression levels present in the remaining population of Jurkat cells compared to the T cells. This would have led to more comprehensive understanding of molecular pathways at play with MR1 interaction. Similarly, the same logic applies to Figure 8.
Future directions for the paper:
The researchers cloned a T cell that specifically did not recognize MHC-1. Therefore, it may be possible to find other types of T cells that can target and kill cancer cells other than MC.7.G5. This could be a start to another type of personalized medicine by co-culture of patient T cells to cancer cells and transducing reactive T cells back.