1.3 Structural cells are key regulators of organ-specific immune responses
DOI: https://doi.org/10.1038/s41586-020-2424-4
Thomas Krausgruber, Nikolaus Fortelny, Victoria Fife-Gernedl, Martin Senekowitsch, Linda C. Schuster, Alexander Lercher, Amelie Nemc, Christian Schmidl, André F. Rendeiro, Andreas Bergthaler & Christoph Bock
Year: 2020
Affiliation CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences, Vienna, Austria
Fig. 1: Multi-omics profiling establishes cell-type-specific and organ-specific characteristics of structural cells.
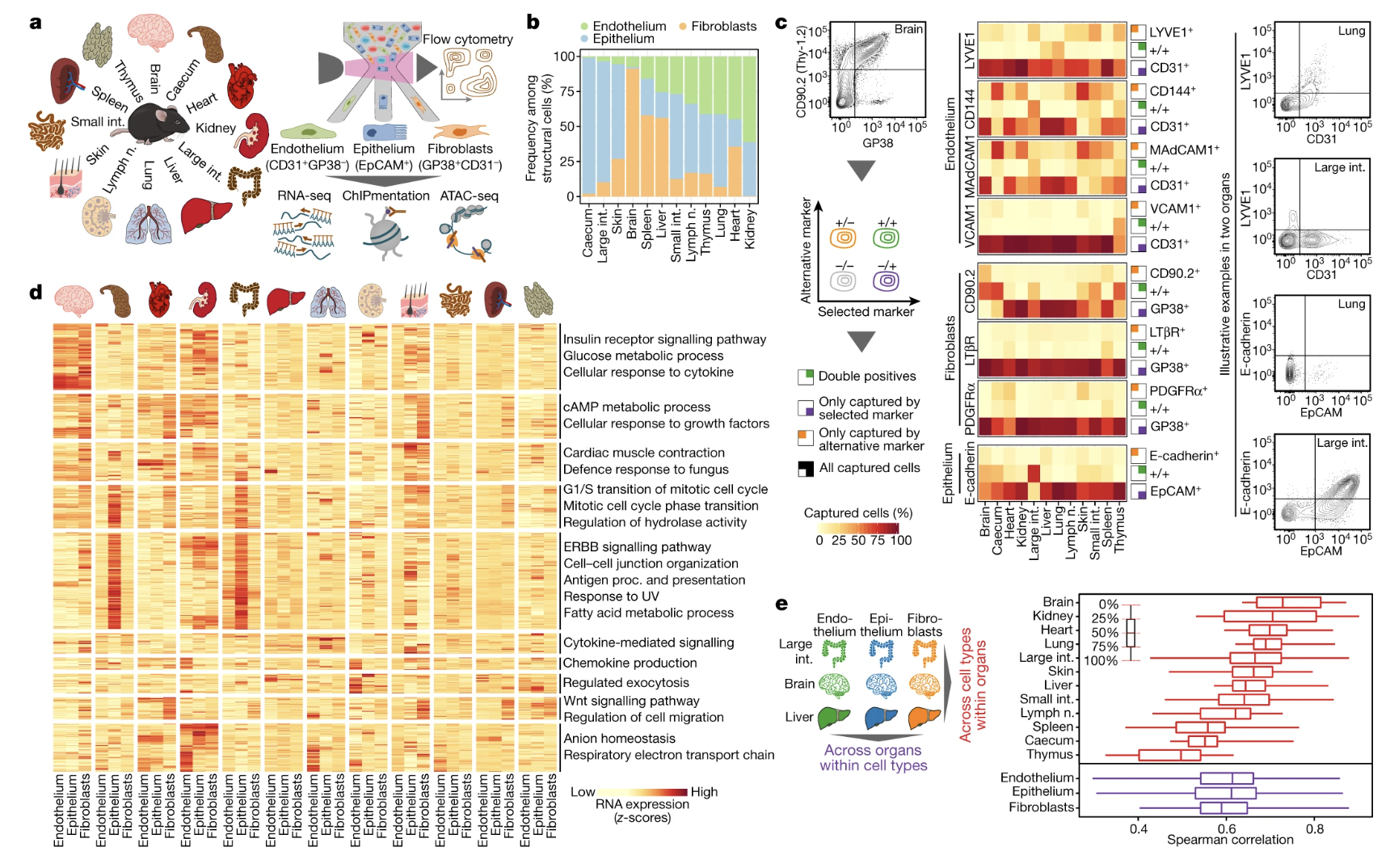
Fig. 2: Gene expression of structural cells predicts cell-type-specific and organ-specific crosstalk with haematopoietic immune cells.
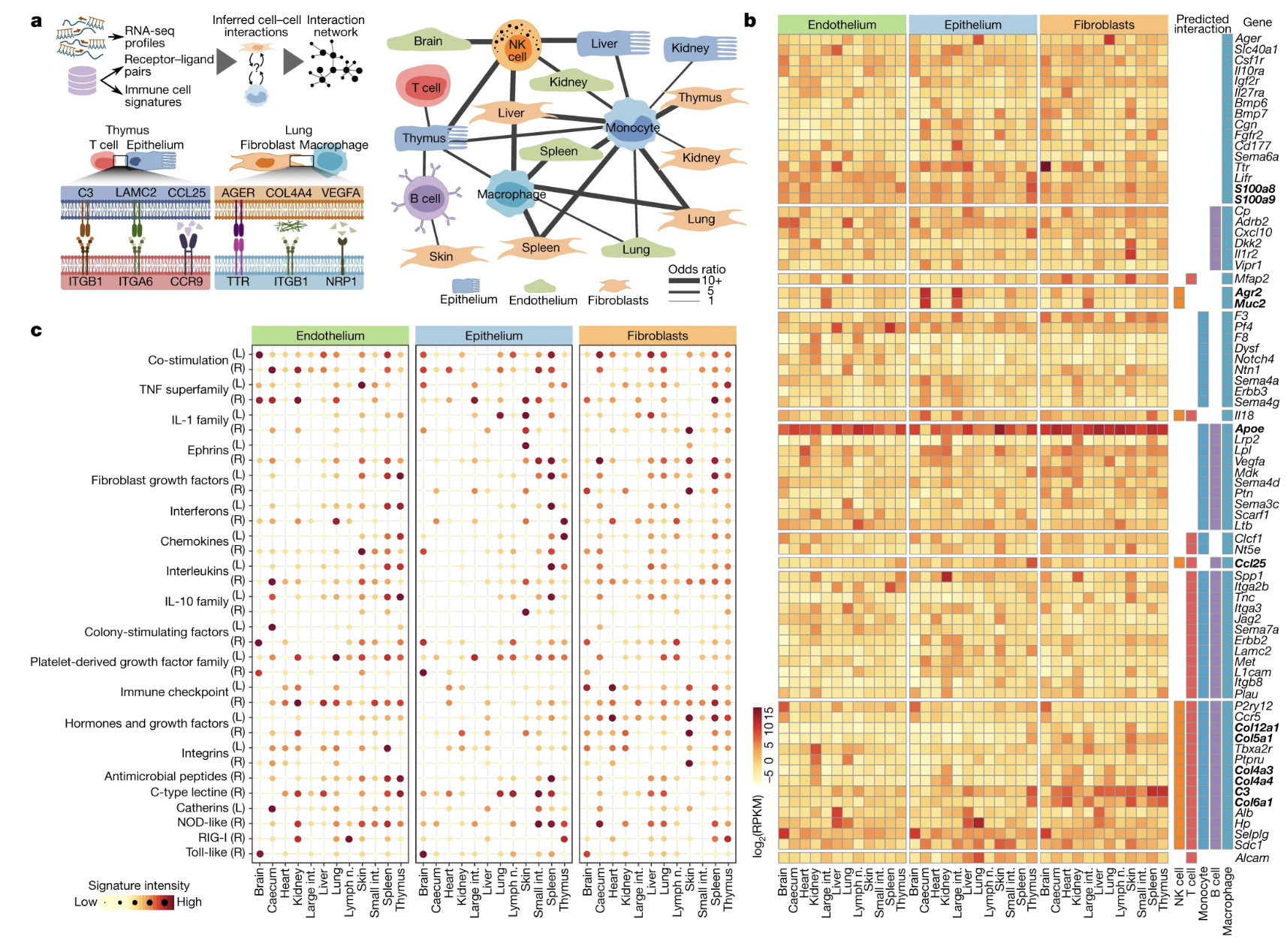
Fig. 3: Structural cells implement characteristic gene-regulatory networks and an epigenetic potential for immune gene activation.
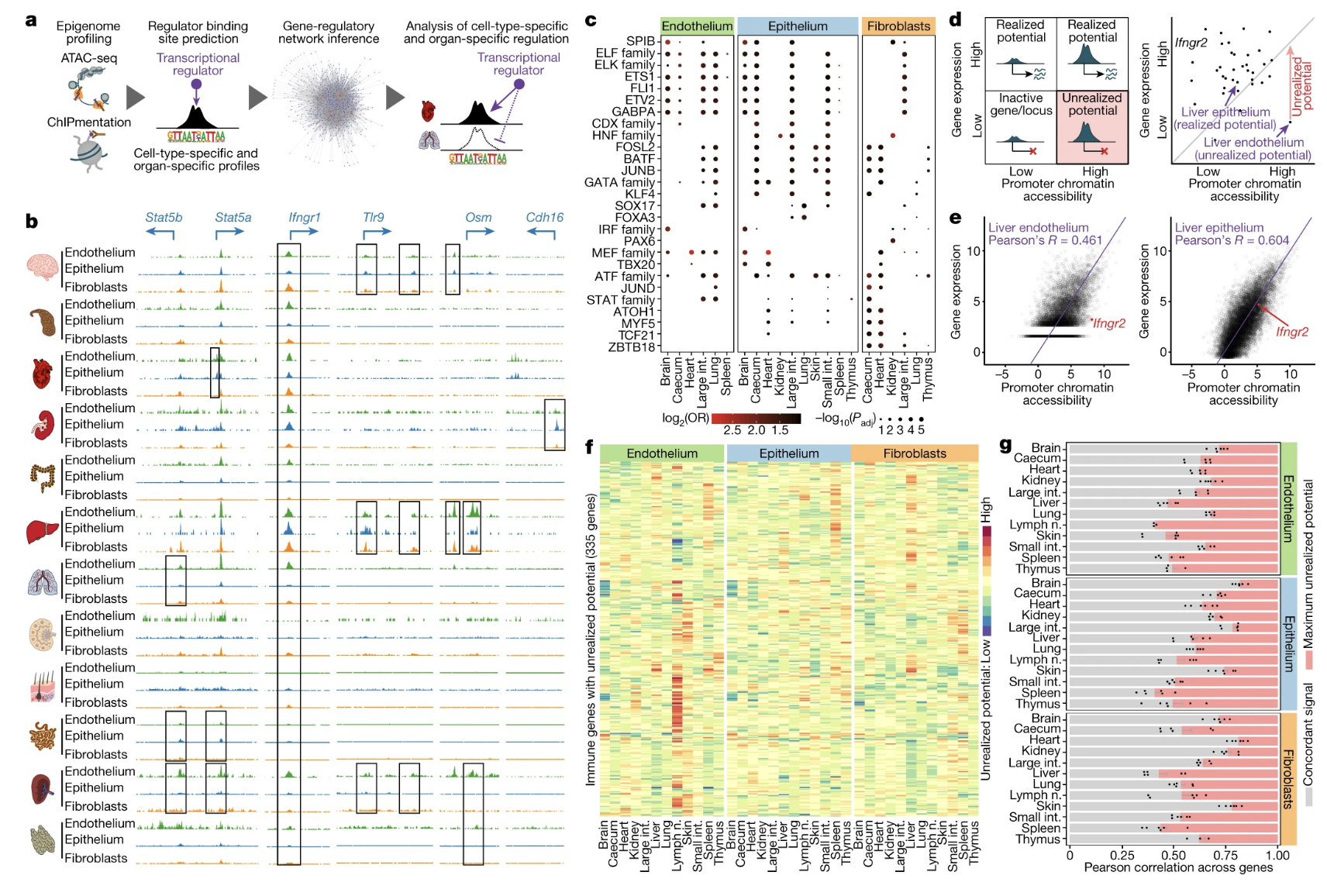
Fig. 4: Systemic viral infection activates the immunological potential of structural cells in vivo.
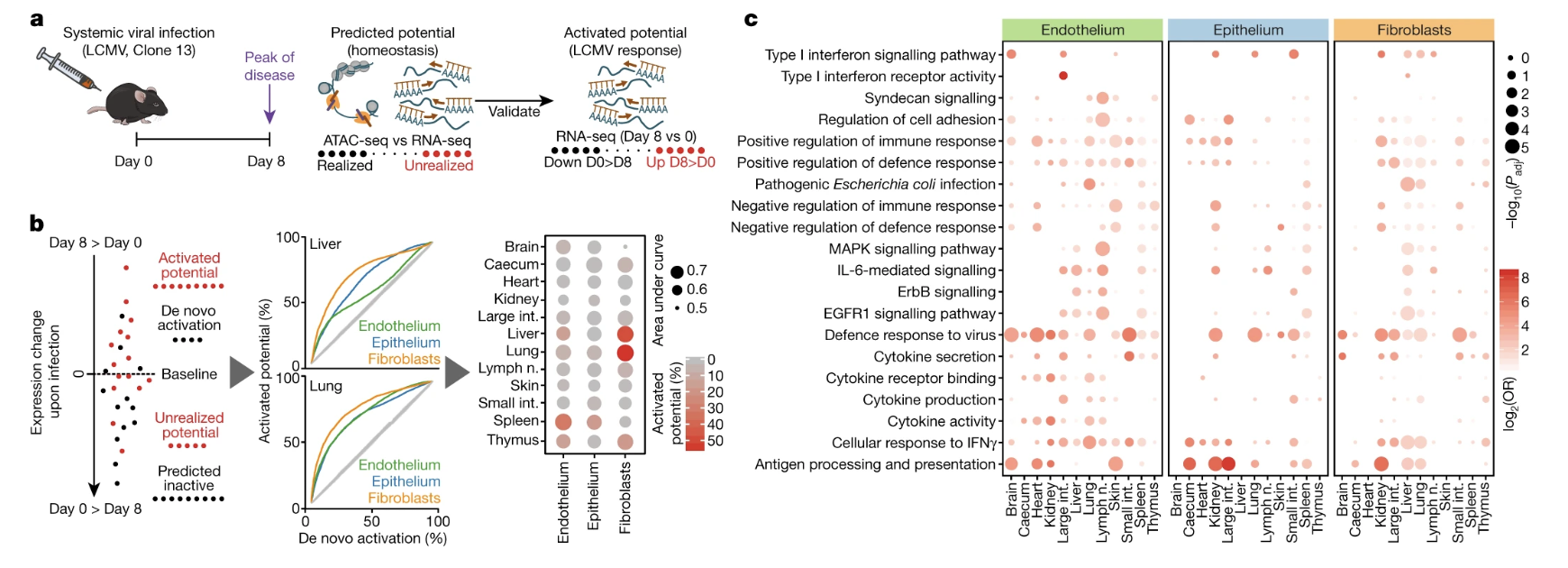
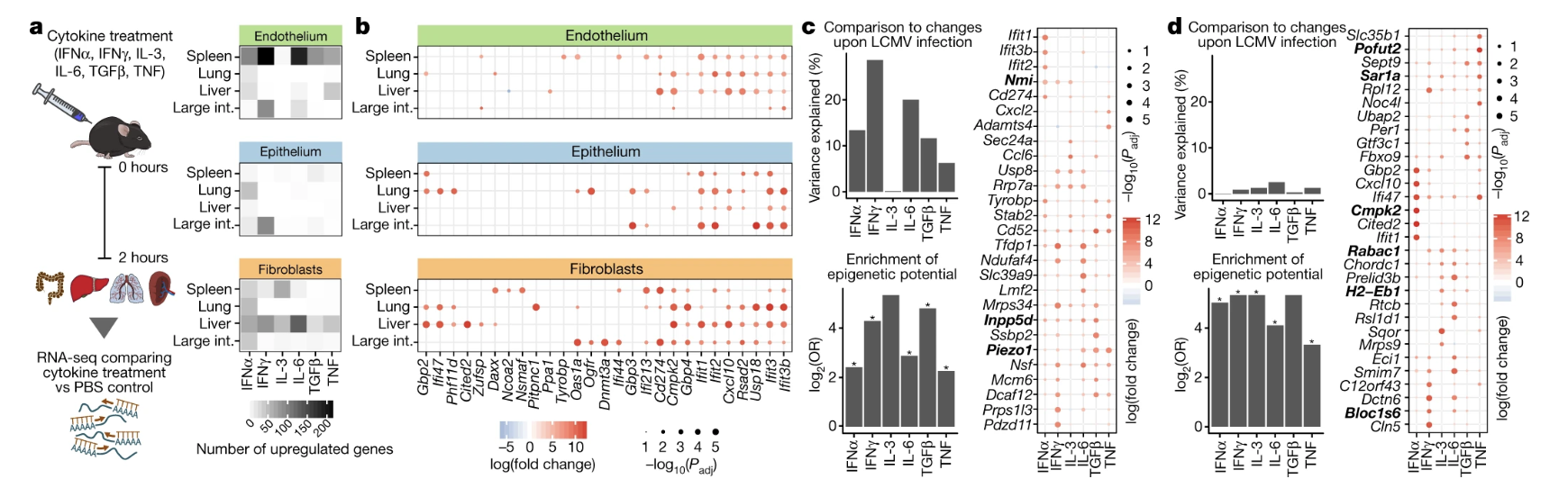
Fig. 5: Cytokine treatment induces cell-type-specific and organ-specific changes in structural cells in vivo.
Main point:
- The researchers aim to unveil the role of structural cells in immune regulation.
- Structural cells are organ-specific and they found structural cells within an organ are more similar to the residing organ than other comparisons.
- This is a bioinformatics heavy paper.
Figures: 1. Details the methodology used to analyze multi-omics profiling including types of sequencing data obtained, cell surface markers, and gating strategy. 2. Characterization of cell to cell interactions based on cell surface receptors that may mediate contact through RNA-seq. Profiles were built to visually depict the receptor and ligands that occur in structural cells, as well as, the genes expressed. 3. Investigates the chromatin structure and potential for epigenetic potential. 4. Tests the model that the researchers built up to this point by infecting a mouse with LCMV 5.
Most Important Figures:
- Figure 4 is one of the most important figures because it is a simple biological test that puts the theories in the last 3 figures together. The researchers place a lot of bioinformatics information in the first 3 figures to exemplify that there is potential for genes to activate under immunological challenge. This data is supported by RNA-seq, ATAC-seq, and ChIPmenation.
- Figure 5
Holes and future directions:
- Figure 2 felt weak from a biological standpoint. They obtained single cell information on expression profiles, cell-cell interaction, and genes that may play immunological function in structural cells. However, for this figure, they never went back to test it by depletion of part of the function, treatment with a drug, etc. Due to testing such a broad group of cell types and organs, the researchers never dove in to identify why an immune specific interaction may work. Therefore, the broad claim feels unsubstantiated despite the data being obtained through bioinformatics.
- Most of Figure 3 also felt weak for the same reasons stated for figure 3. Additionally, they state that the data is a concrete example of epigenetic potential, but it doesn’t look like they tested this in a biological study. Interestingly, figure 3f felt like the strongest point in the figure that depicts a heat map of genes with potential roles in structural cells.
- The benefit of this paper is that it is more methodical. This appears to set the opportunity to fish for genes of interest to study. This is probably the reason why it is more broad than specific.